Understanding unwarranted variations in emergency care
Benchmarking mortality risk prediction from electrocardiograms
Background
Electrocardiography measures a heart’s electric activity and the resulting plot, an electrocardiogram, can be used to diagnose pathologies. ECGs (an acronym that refers to either the process or the resulting graph, depending on context) are easy to collect, and results are interpreted in a standard manner that primarily focuses on peak widths and heights. The ECG is a time-series signal that usually has 12 channels (although only 8 are linearly independent). As the heart contracts, the resulting movement of ions creates voltage changes that can be measured from the surface of the subject’s skin. Since ECGs are cheap, they are used to screen patients before using more expensive diagnostic tools (like MRIs).
Publicly available ECG datasets have gotten very big since 2018, with recent studies building models from hundreds of thousands to millions of ECGs. One study1 used ECGs from a giant telehealth network in Brazil and predicted people’s ages. A portion of this dataset is available (Code-15) and commonly used in ECG modeling. They found that people who’s predicted ages were higher than their chronological age were at a higher risk of dying in the next few years, and the opposite was true for people who’s predicted ages were lower than their actual age. Since then, many other studies have worked on predicting mortality timelines from ECG measures.
We were curious if similar approaches could be used in Boston Children’s Hospital to stratify patient risk (if the risk is that of mortality, this task is equivalent to mortality prediction). Stratifying patient risk could potentially allow more patient-tailored diagnostic schedules, such as increasing or decreasing the frequency of expensive and cumbersome medical tests (again, MRIs, which in pediatric cases often include patient sedation to minimize movement).
Most past studies used private data from US hospitals and, surprisingly, used many different ECG-interpretation approaches (mostly Convolutional Neural Networks, but also tree-based approaches from machine-extracted features) and survival modeling approaches (usually classifiers predicting a mortality by year h followed by Cox regressions, but sometimes Deep Survival models that generate the resulting curve directly). While performance was generally similar (AUROC ~0.8) and sometimes models were evaluated on outside datasets, the models typically weren’t compared to simpler baselines.
No past work had systematically compared these modeling approaches, so that’s where this project started. We used three datasets: MIMIC-IV ECG (785k ECGs)2, which had recently come out, Code-15 (233k), and a dataset from Boston Children’s Hospital (BCH, 182k)3.
Modeling
We used two ECG-interpreting approaches, the ResNet structure from Ribeiro and InceptionTime4, which our group had recently used pretty successfully in fetal heart-rate monitoring5, and eight survival analysis approaches – four Classifier-Cox models built off classifiers predicting death by 1,2,5,or 10-years, and four Deep Survival approaches from the PyCox package6.
Models were trained with and without demographics (age and sex), and in MIMIC’s case we also tried using automatic machine measurement values (like QRS duration). The covariates were processed with a three-layer 32-dim MLP, and those results were fused with CNN outputs using a 3-layer, 128-dimension MLP.
We also trained the models with baselines that did not directly look at the ECG signal: XGB Classifier-Cox models (in MIMIC’s case, these could look at the machine measures) and the previously described architecture but with the ‘CNN’ portion returning a constant ‘0’ (so effectively a 6-layer feedforward model).
So, at least in MIMIC’s case, we included both 20-year-old approaches (ECG Features -> XGB/MLP -> Cox) and modern approaches (Raw ECG -> CNN tailored to ECG architectures -> Deep Survival).
Results
This is the key figure from the results:
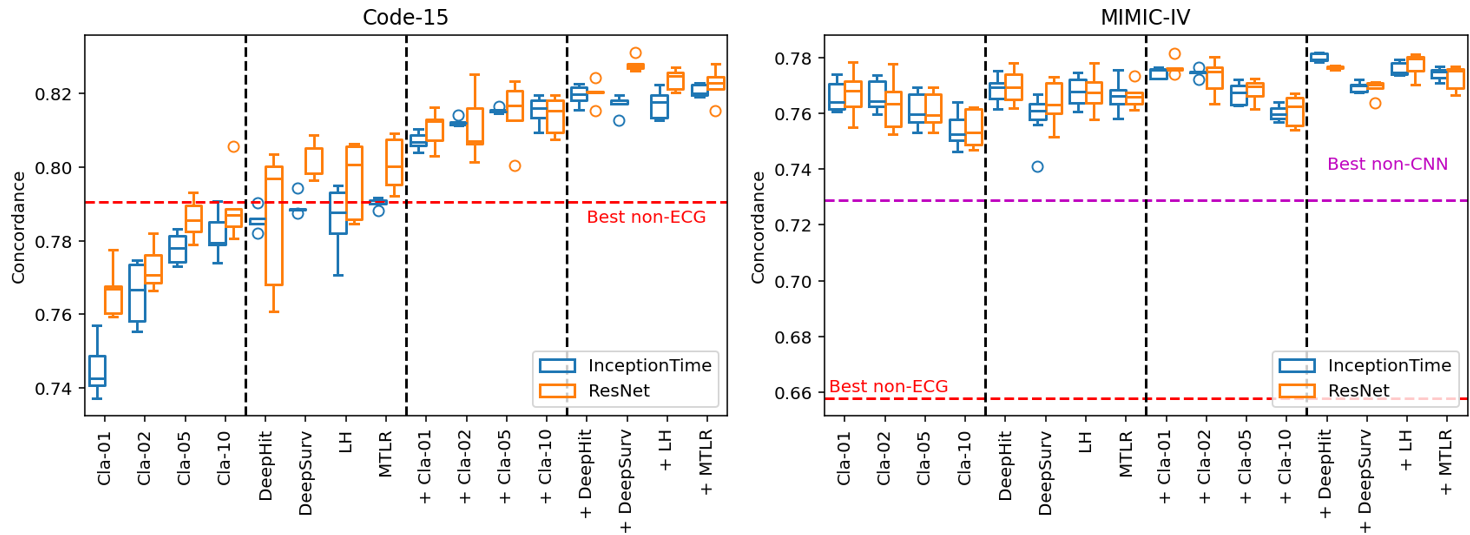
There are four main conclusions:
- Demographic-only (non-ECG) models can be surprisingly competitive (Code-15 red line); models should be compared to these rather than the 0.5 “chance” rate.
- Including demographics in ECG models improves AI-ECG performance (for Code-15, enough to consistently beat the non-ECG models)
- Deep-Survival models are generally better than Classifier-Cox models.
- Classifier-Cox models are quite sensitive to how you define the prediction horizon (i.e., predicting mortality in one year versus five). Deep-Survival models handle time horizons more elegantly, by design.
We also trained BCH models with the best-performing approach (ResNet LogisticHazard), and cross-evaluated all models of this type. Performance can drop substantially on cross-evaluation, sometimes falling far below the performance of demographic-only models (BCH and MIMIC models score 0.54 and 0.70 on Code-15’s test set, vs Code-15’s demographic-only 0.79).
Discussion
Systematically sweeping model configurations suggests that deep survival approaches should be used (better performance, fewer parameters to tune), that demographics (age/sex) contribute to accuracy, and that simpler models (e.g. demographic-only) should be the comparative baseline when evaluating AI-ECG models. This would improve model performance and clarify the value provided by AI-ECG models.
Futher Reading
For more details and open-source code for benchmarking AI-ECG models, see below:
References
-
Antônio H. Ribeiro, Manoel Horta Ribeiro, Gabriela M. M. Paixão, Derick M. Oliveira, Paulo R. Gomes, Jéssica A. Canazart, Milton P. S. Ferreira, Carl R. Andersson, Peter W. Macfarlane, Wagner Meira, Thomas B. Schön, and Antonio Luiz P. Ribeiro. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nature Communications, 11(1):1760, April 2020 ↩
-
A. L. Goldberger, L. A. N. Amaral, L. Glass, J. M. Hausdorff, P. Ch. Ivanov, R. G. Mark, J. E. Mietus, G. B. Moody, C.-K. Peng, and H. E. Stanley. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation, 101(23):e215–e220, 2000 (June 13). ↩
-
Hassan Ismail Fawaz, Benjamin Lucas, Germain Forestier, Charlotte Pelletier, Daniel F. Schmidt, Jonathan Weber, Geoffrey I. Webb, Lhassane Idoumghar, Pierre-Alain Muller, and François Petitjean. InceptionTime: Finding AlexNet for Time Series Classification. Data Mining and Knowledge Discovery, 34(6):1936–1962, November 2020. arXiv:1909.04939[cs, stat] ↩
-
Håvard Kvamme, Ørnulf Borgan, and Ida Scheel. Time-to-Event Prediction with Neural Networks and Cox Regression. Journal of Machine Learning Research, 20(129):1–30, 2019. ↩
2025
Taking a closer look at survival modeling with ECGs
2024
Relaxing the definition of equivalent mathematical expressions to get simpler and more interpretable models
2023
About our recent HUMIES award-winning algorithm for clinical prediction models
A new perspective on how this social theory relates to fair machine learning.
2022
We consistently observe lexicase selection running times that are much lower than its worst-case bound of \(O(NC)\). Why?
